In this experiment we are looking at two strains of Arabidopsis thaliana. One is the wild type which is highly susceptible to infection by Sclerotinia and the other is a mutant, which is significantly more resistant. We are looking for a set of genes which are expressed differentially in the two strains.
6 plants of each strain were infected with Sclerotinia. After 4 hours, 3 individuals of each strain were ‘harvested’ and RNA was extracted from the leaf tissue. The remaining samples were harvested after 14 hours and prepared in the same way.
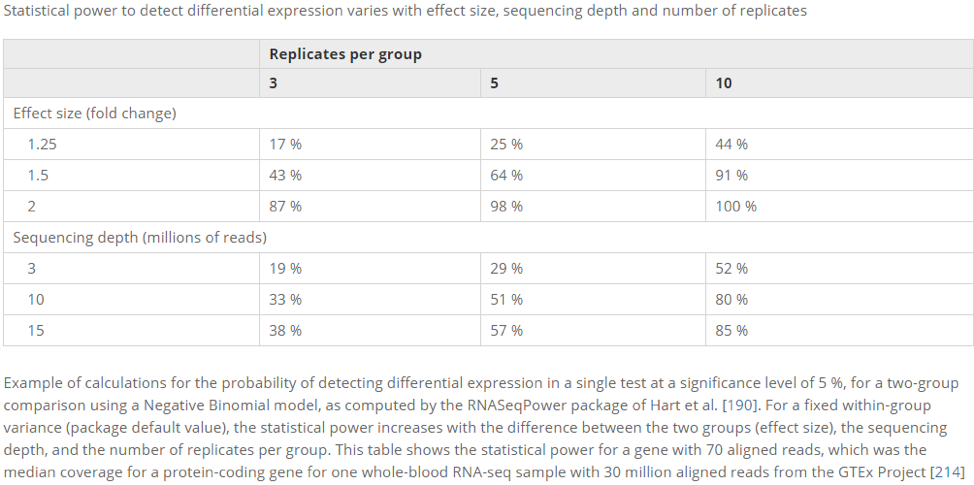
The importance of replicates cannot be understated. Without sufficient replicates it will be difficult to distinguish a real pattern of expression between conditions from background biological variation present within conditions.
The cost of this replication can be significant, both in terms of the experimental time and effort required and also because the majority of the cost of sequencing experiments comes from the library preparation required prior to sequencing. However, they are absolutely critical.
Three replicates are considered the absolute minimum for each condition. Even with three replicates you run the risk is that if one sample does not yield good sequence the other two in the set will be insufficient to allow a calculation of variance.. Even with 3 replicates you can expect a good number of false-positives and the ability to detect smaller effects is diminished.
Imagine if you are running a project to determine if men are taller or shorter than women on average - you decide to sample 3 of each gender - how confident would you be about your findings? Now consider you are doing this tens of thousands of times (once for each gene) how many might yield misleading results just by chance.
The table below shows the consequence of replicate numbers on the ability to detect differences at various fold change levels. For example if have 3 replicates you will have 43% chance of detecting a gene with a 1.5x fold change.